CANDESIGN
Computational ANalytics to DESIGN

Overview
CANDESIGN is a tool for differential target based design that utilizes the docking functionality provided by CANDOCK. It allows for automated drug design to take place after docking has occurred via CANDOCK using the fragments generated with CANDOCK.
How it works
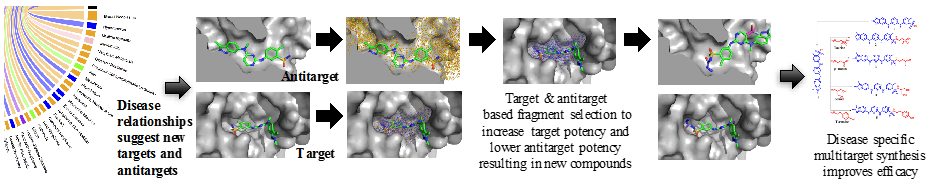
CANDESIGN is a software toolkit based on the features developed in CANDOCK. The user has an option to give a starting lead molecule or let the program design a new scaffold de novo and, if given, the lead compounds are docked as described on the CANDOCK page. This docking is carried out along with additional 'design' fragments supplied by the user using the dock_fragment tool in the CANDOCK software package. These design fragments can be independent molecules or fragments from a larger database (such as all FDA approved drugs). These leads (if given) and fragments are docked to a list of targets and antitargets, also provided by the user.
Once the docking has completed, CANDESIGN selects high scoring fragments in the targets that are low scoring in the antitargets. It then attaches these fragments to the leads and re-docks the resulting molecule. This new molecule undergoes the same design process until the resulting compound violates a set of rules governed by the user (for example, the user may wish to create compounds that must obey Lipinski's Ro5).
Examples

Tutorial
cp /depot/gchopra-class/data/additional.mol2 .
cp /depot/gchopra-class/data/lead.mol2 .
mkdir targets
cp /depot/gchopra-class/data/target.pdb targets/
mkdir atargets
cp /depot/gchopra-class/data/antitarget.pdb atargets/
export CANDOCK_ligand=lead.mol2
export CANDOCK_fragment_mol=additional.mol2
export CANDOCK_target_dir=targets
export CANDOCK_antitarget_dir=atargets
submit_candock_module.sh design_ligands -q scholar -l walltime=30:00:00