Purdue publications
(2016 - Present)

Identification of monomethyl branched chain lipids by a combination of liquid chromatography tandem mass spectrometry and charge-switching chemistries
Randolph C, Beveridge C, Iyer S, Blanksby S, McLuckey S, Chopra G†
JASMS (2022)
[JASMS]


[ChemRxiv]

[ChemRxiv]

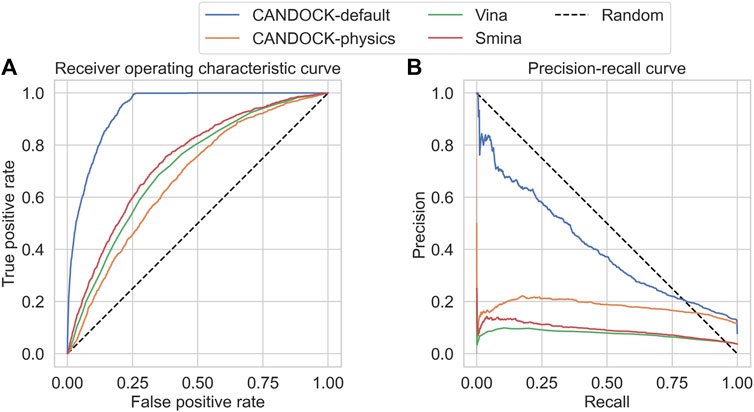
Accurate Prediction of Inhibitor Binding to HIV-1 Protease using CANDOCK
Falls Z, Fine J, Chopra G, Samudrala R.
Fronteirs in Chemistry (2021)


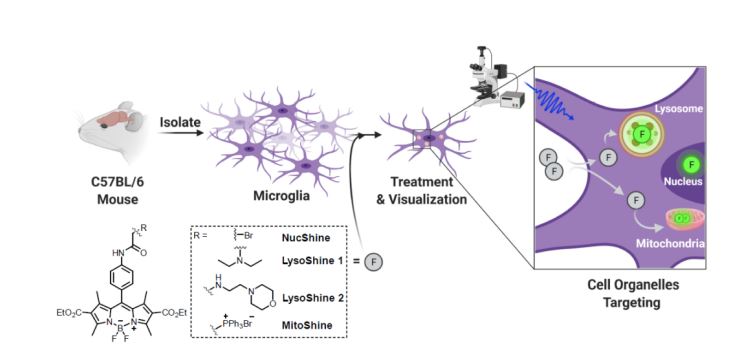
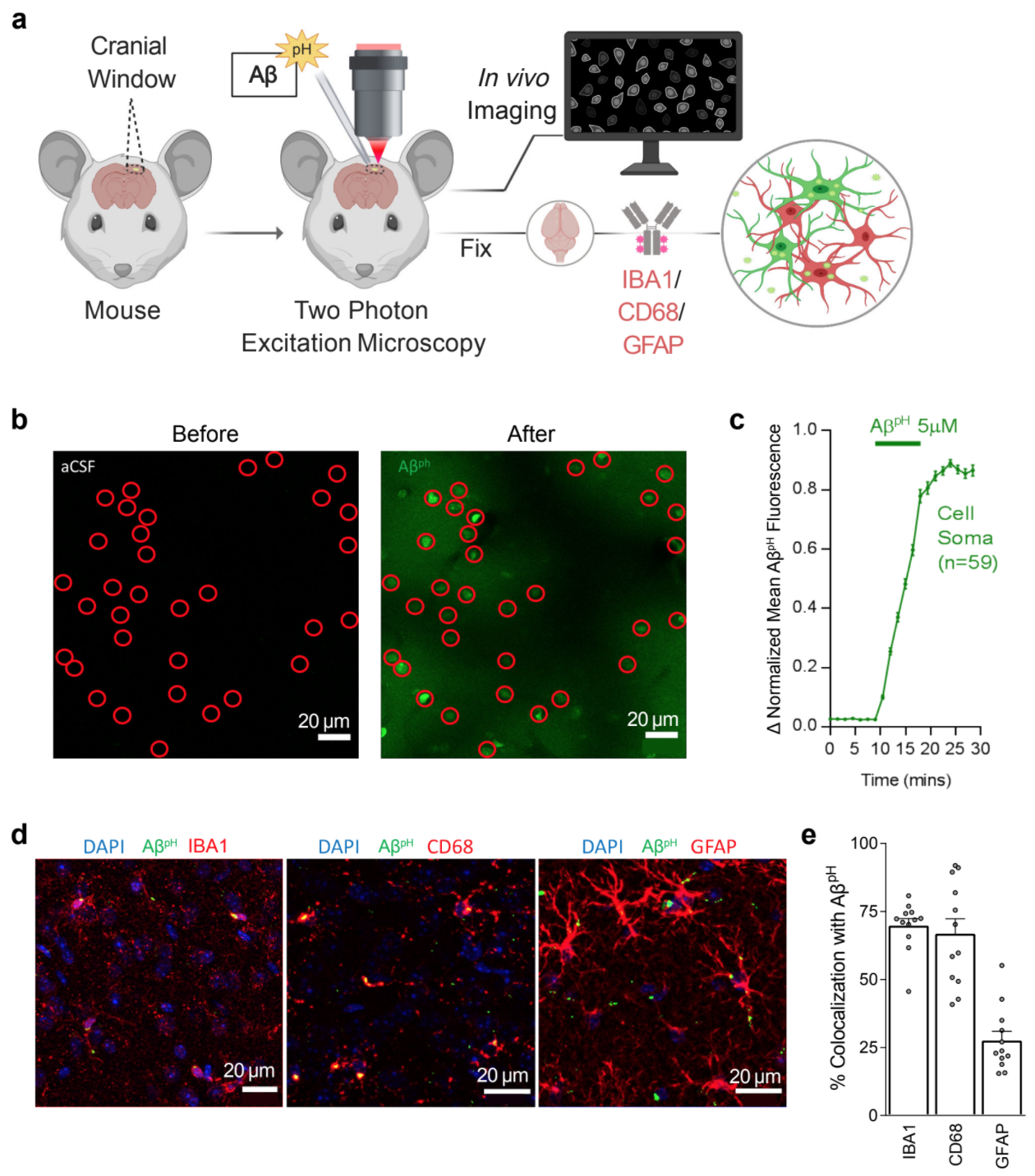
Monitoring phagocytic uptake of amyloid β into glial cell lysosomes in real time
Prakash P*, Jethava KP*, Korte N*, Izquierdo P*, Favuzzi E, Rose IVL, Guttenplan KA, Manchanda P, Dutta S, Rochet J-C, Fishell G, Liddelow S, Attwell D, Chopra G†

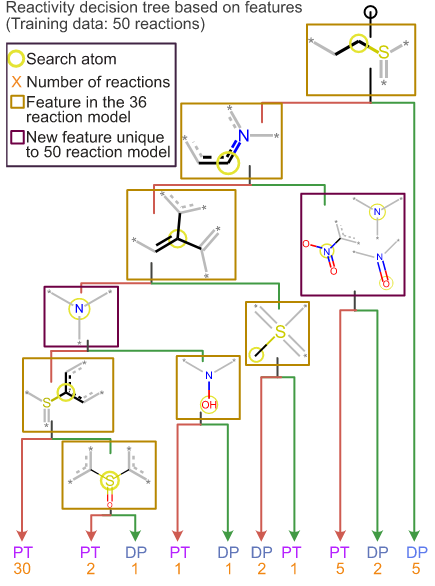
Graph based machine learning interprets and predicts diagnostic isomer-selective ion-molecule reactions in tandem mass spectrometry.
Fine J, Li JK-Y*, Beck A, Alzarieni K, Ma X, Boulos V, Kenttämaa HI, Chopra G†
Chem. Sci. 11, 11849-11858 (2020).

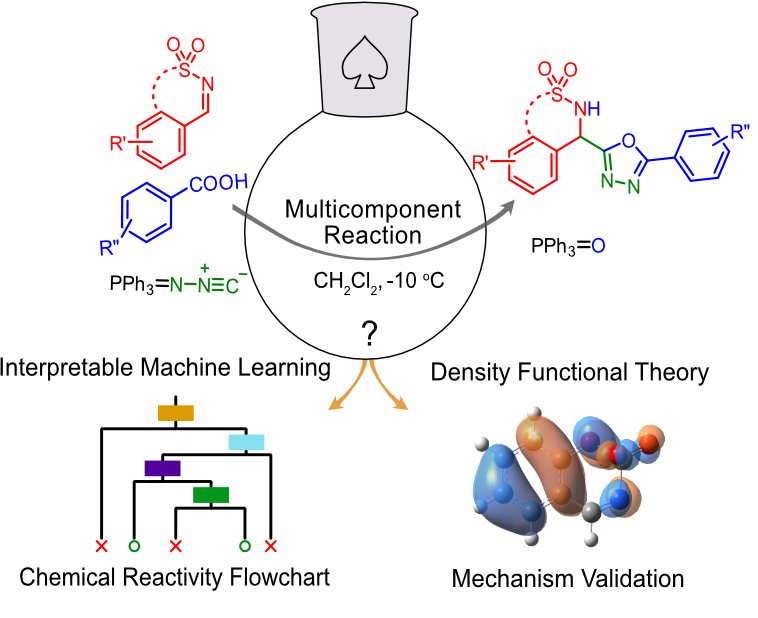
Accelerated reactivity mechanism and interpretable machine learning model of N-Sulfonylimines towards fast multicomponent reactions
Jethava KP, Fine JA, Chen Y, Hossain A, Chopra G†
Org. Lett. 22, 21, 8480–8486 (2020).

A 2‐Tyr‐1‐Carboxylate Mononuclear Iron Center is the Active Site of Dimethylformamidase
Arya CK, Yadav S, Fine J, Casanal A, Chopra G†, Ramanathan G, Vinothkumar KR, Subramanian R†.

Spectral Deep Learning for Prediction and Prospective Validation of Functional Groups
Fine J, Jethava K,
Chem. Sci. 11, 4618-4630 (2020).
Targeting polyamine biosynthesis to stimulate beta cell regeneration in zebrafish
Robertson M ,Padgett L, Fine J, Chopra G, Mastracci T †
Islets. Sep 2;12(5):99-107 (2020).
[ISLETS]

Mangione W, Falls Z, Chopra G, Samudrala R†
[JCIM]

DUBS: A framework for developing Directory of Useful Benchmarking Sets for virtual screening.
Fine J, Muhoberac M, Fraux G, Chopra G†
J. Chem. Inf. Model. 60, 9, 4137-4143 (2020).
[JCIM]

Shotgun Drug Repurposing Biotechnology to Tackle Epidemics and Pandemics
Mangione W, Falls Z, Melendy T, Chopra G†, Samudrala R†

CANDOCK: Chemical atomic network based hierarchical flexible docking algorithm using generalized statistical potentials
Fine J, Konc J, Samudrala R, Chopra G†
J. Chem. Inf. Model. 2020, 60, 3, 1509-1527 (2020).
[JCIM]

[BioRxiv]

Graph Neural Networks Bootstrapped for Synthetic Selection and Validation of Small Molecule Immunomodulators
Wijewardhane PR, Jethava KP, Fine JA, Chopra G†
[ChemRxiv]

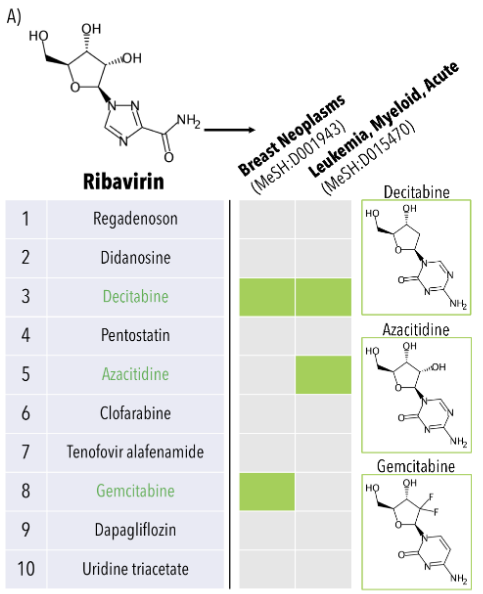
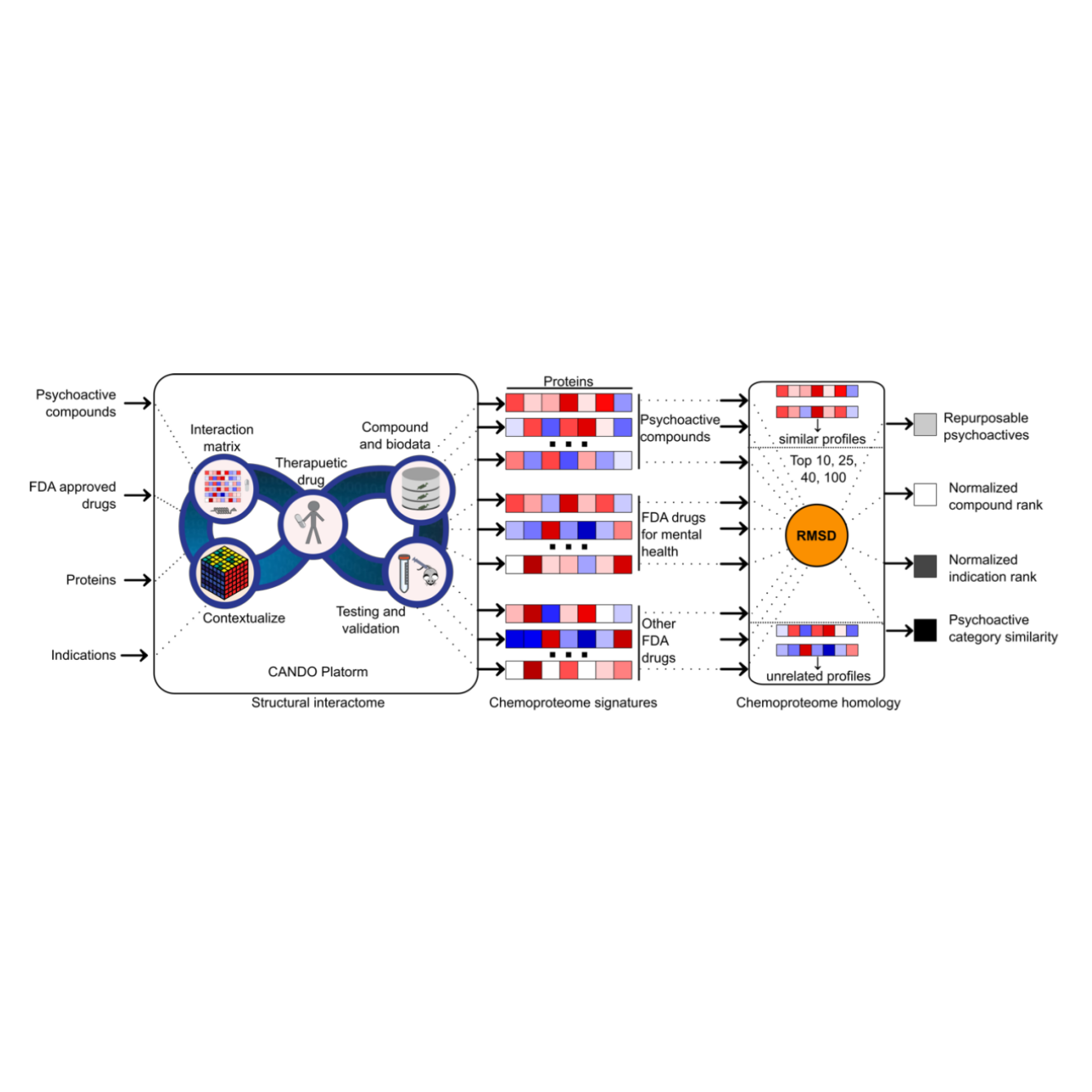
Computational chemoproteomics to understand the role of selected psychoactives in treating mental health indications
Fine J, Lackner R, Samudrala R, Chopra G†
Scientific Reports, 9:13155 (2019).

Integrated pan-cancer map of EBV-associated neoplasms reveals functional host-virus interactions
Chakravorty S, Yan B, Wang C, Wang L, Quaid JT, Lin CF, Briggs SD, Majumder J, Canaria DA, Chauss D, Chopra G, Olson MR, Zhao B, Afzali B, Kazemian M

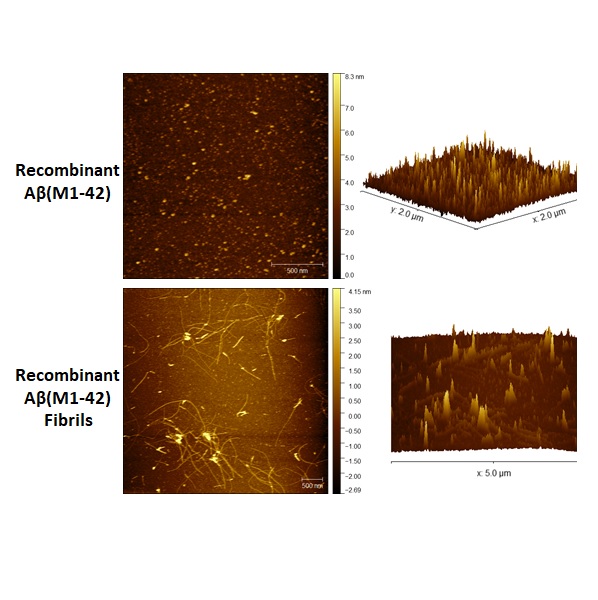
Rapid, Refined, and Robust Method for Expression, Purification, and Characterization of Recombinant Human Amyloid-beta M1-42
Prakash P, Lantz TC, Jethava K, Chopra G†
Methods Protoc. 2019, 2(2), 48 (2019).

Lemon: a framework for rapidly mining structural information from the Protein Data Bank
Fine J, Chopra G†
Bioinformatics, 35(20): 4165–4167 (2019).


Hossain A, Majumder J, Bi C, Huang F, Chopra G†

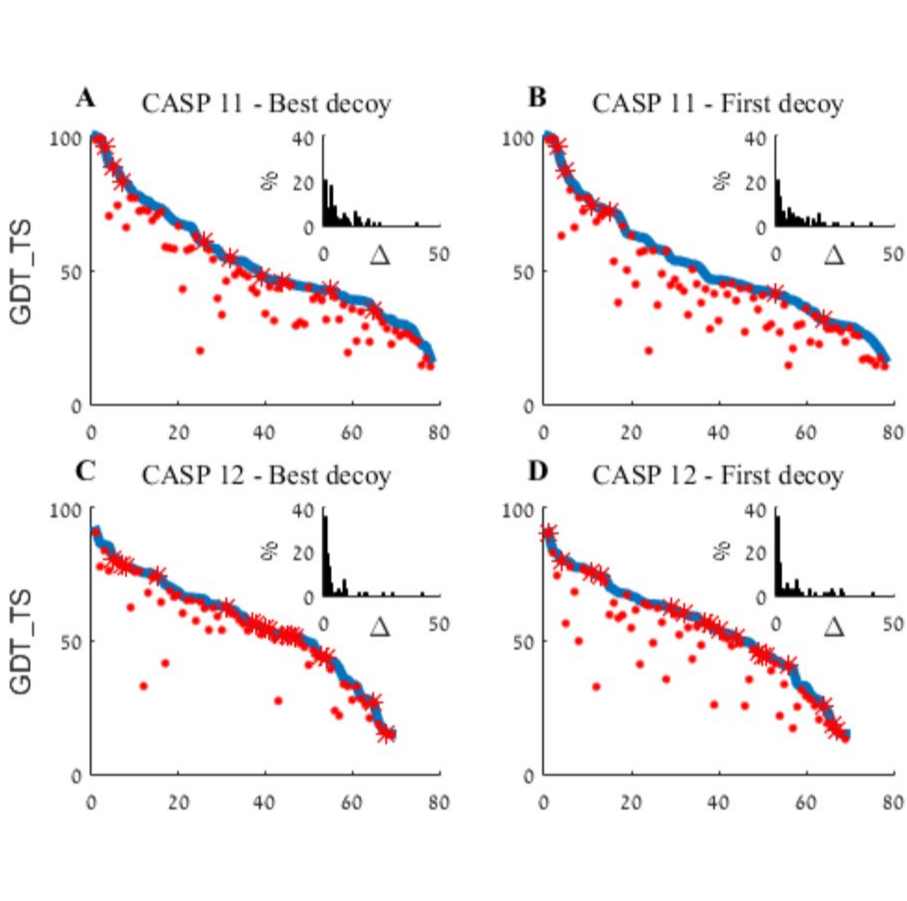
An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12
Keasar C, McGuffin LJ, Wallner B, Chopra G, et. al.
Scientific Reports, 2018, 8:9939 (2018).
Inhibition of 12/15-lipoxygenase protects against ß cell oxidative stress and glycemic deterioration in mouse models of type 1 diabetes.
Hernandez-Perez M, Chopra G, Fine J, Conteh AM, Anderson RM, Linnemann AK, Benjamin C,
Nelson JB, Benninger KS, Nadler JL, Maloney DJ, Tersey SA, Mirmira RG.
Diabetes, 2017, [Epub ahead of print]
PMID: 28842399

Aminoisoquinoline benzamides, FLT3 and Src-family kinase inhibitors, potently inhibit proliferation of acute myeloid leukemia cell lines
Larocque E, Naganna N, Ma X, Opoku-Temeng C, Carter-Cooper B, Chopra G, Lapidus RG, Sintim HO.
Future Med Chem, 2017, 9(11), 1213-1225
PMID:28490193

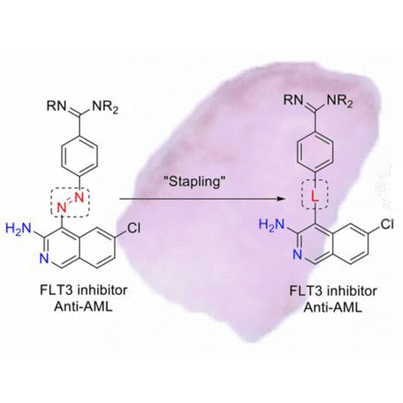
Identification of new FLT3 inhibitors that potently inhibit AML cell lines, via an azo click-it/staple-it approach
Zhou J, Ma X, Wang C, Carter-Cooper B, Larocque, E, Fine J,
Tsuji G, Yang F, Chopra G, Lapidus RG, Sintim HO*.
ACS Med Chem Lett, 2017, 8 (5), 492–497
PMID: 28523099

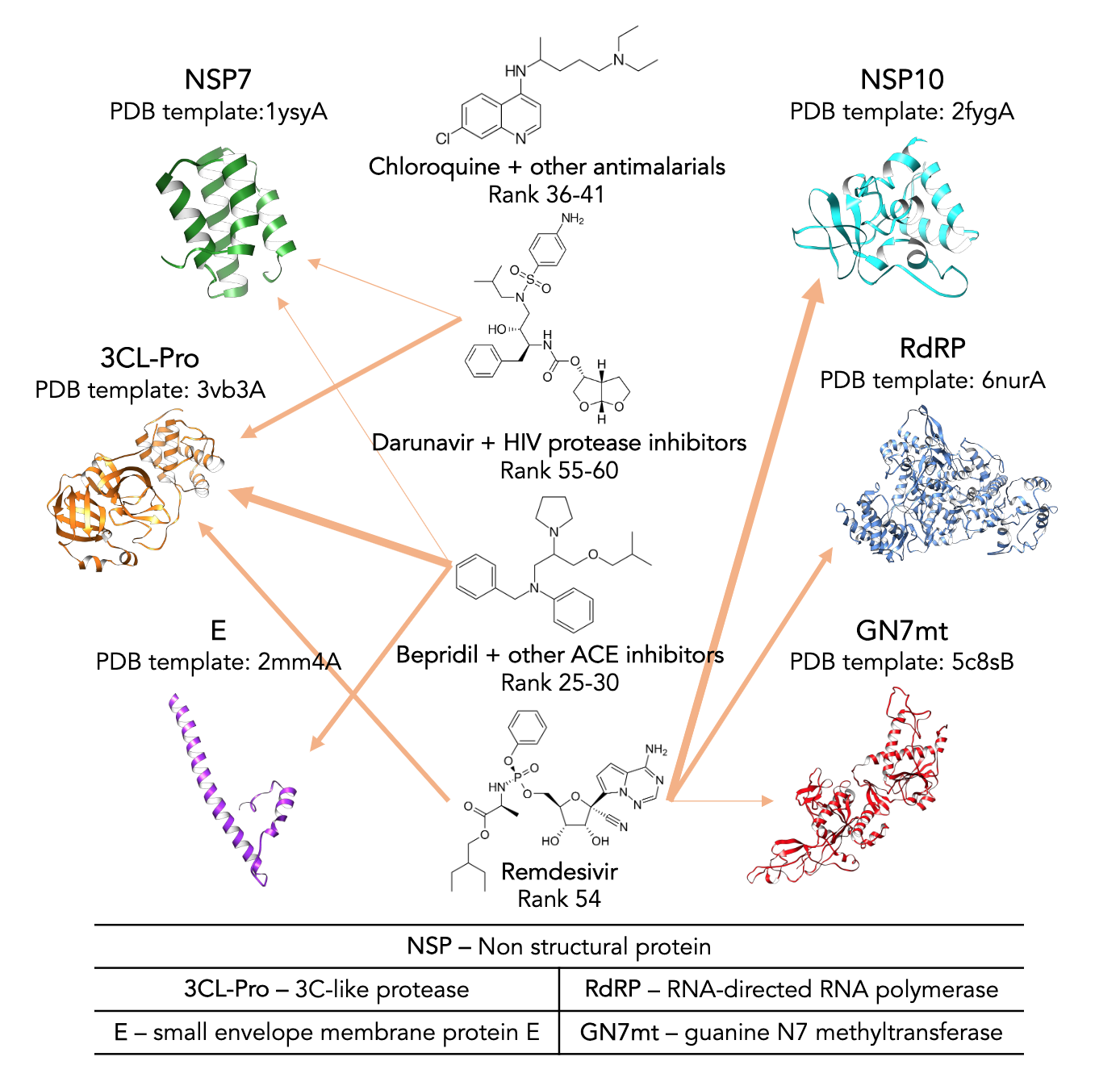
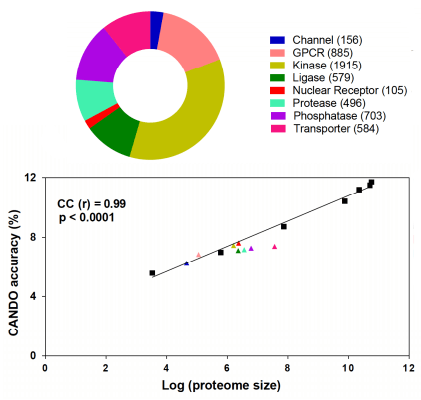
Combating Ebola with Repurposed Therapeutics Using the CANDO Platform
Chopra G†, Kaushik S, Elkin PL, Samudrala R
Molecules, 2016, 21(12): 1537
PMID:27898018
Summary: With the use of the CANDO platform and in vitro data, a significant reduction in time, risk, cost, and resources was shown when identifying drug leads for future Ebola outbreaks compared to tradition methods.

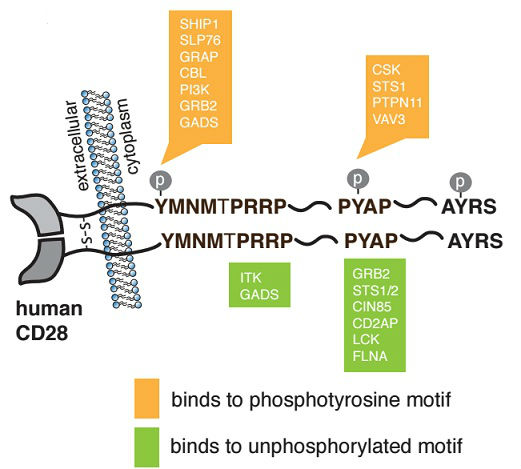
CD28 Costimulation: From Mechanism to Therapy
Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA
Immunity, 2016, 44(5): 973-988
PMID: 27192564
Summary: This paper summarizes the current understanding of complex costimulatory pathways, including the individual roles of the CD28, B7-1/CD80, and B7-2/CD86 molecules.

Exploring polypharmacology in drug discovery and repurposing using the CANDO platform
Chopra G†, Samudrala R
Current Pharmaceutical Design, 2016, 22(21):3109-23
PMID: 27013226
Summary: Shows how the CANDO platform can be used to analyze and determine the contribution of
Stanford and UCSF publications
(2008 - 2015)

Divergent phenotypes of human regulatory T cells expressing the receptors TIGIT and CD226
Fuhrman CA, Yeh W-I, Seay HR, Lakshmi PS, Chopra G, Zhang L, Perry DJ, McClymont SA, Yadav M, Lopez MC, Baker HV, Zhang Y, Li Y, Whitley M, von Schack D, Atkinson MA, Bluestone JA, Brusko TM
Immunology, 2015, 195(1): 145-155
PMID: 25994968
Summary: A set of studies elucidating functionally distinct regulatory T-cell subsets that may help direct cellular therapies and provide important phenotypic markers for assessing the role of Tregs in health and disease.

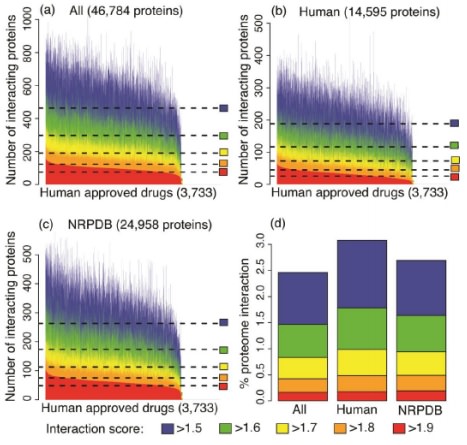
Multiscale modeling of relationships between protein classes and drug behavior across all diseases using the CANDO platform
Sethi G*, Chopra G*, Samudrala R
Medicinal Chemistry, 2015, 15(8): 705-17
PMID: 25694071
Summary: Uses the CANDO platform to analyze the interaction signatures between human approved compounds and the druggable proteome to determine drug behavior for different indications/diseases.

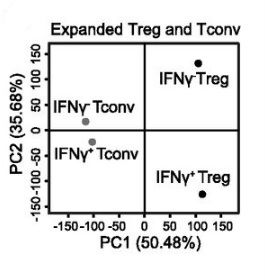
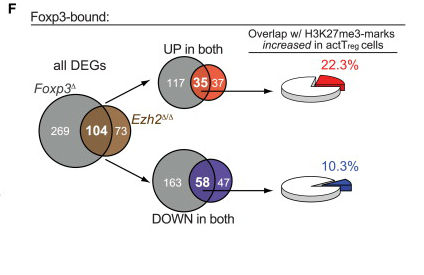
The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T-cell identity
DuPage M, Chopra G, Quiros J, Rosenthal WL, Morar MM, Holohan D, Zhang R, Turka L, Marson A, Bluestone JA
Immunity, 2015, 42(2): 227-38
PMID: 25680271
Summary: Chromatin-modifying enzyme along with CD28 costimulation are crucial for the maintenance of Treg cell identity during cellular activation.

Oxido-reductive regulation of human vascular remodeling by the orphan receptor tyrosine kinase ROS1
Ali Za, de Jesus Perez V, Yuan K, Orcholski M, Pan S, Qi W, Chopra G, Adams C, Kojima Y, Leeper NJ, Qu X, Zaleta-Rivera K, Kato K, Yamada Y, Oguri M, Kuchinsky A, Channon K, Charest A, Quertermous T, Ashley EA
J. Clinical Investigation, 2014, 124(12): 5159-74
PMID: 25401476
Summary: This study indicates that glutathione peroxidase-1 (GPX1)-dependent alterations in

CANDO and the infinite drug discovery frontier
Minie ME*, Chopra G*, Sethi G, Horst JA, Roy A, White G, Samudrala R
Drug Discovery Today, 2014, 19(9):1353-63
PMID: 24980786
[PDF]Summary: The Computational Analysis of Novel Drug Opportunities (CANDO) platform (http://protinfo.org/cando) uses similarity of compound-proteome interaction signatures to infer homology of compound/drug behavior.

WeFold: Large-scale Coopetition for Protein Structure Prediction
Khoury GA, Liwo A, Khatib F, Zhou H, Chopra G, Bacardit J, Bortot LO, et. al.
Proteins, 2014, 82(9):1850-1868
PMC:4249725
Summary: This manuscript describes both successes and areas needing improvement as identified throughout the first WeFold experiment and discusses the efforts that are underway to advance this initiative.

Distal effect of amino acid substitutions in CYP2C9 polymorphic variants causes differences in interatomic interactions against (S)-warfarin
Lertkiatmongkol P, Assawamakin A, White G, Chopra G, Rongnoparut P, Samudrala R, Tongsima S
PLoS One, 2013, 8(9): e74053
PMID: 24023924

Strategic protein target analysis for developing drugs to stop dental caries
Horst JA, Pieper U, Sali A, Zhan L, Chopra G, Samudrala R, Featherstone JDB
Adv. Dent. Res., 2012, 24(2): 86-93
PMID: 22899687
Summary: The entire Streptococcus

KOBAMIN – KnOwledge BAsed MINimization server for protein structure refinement
Rodrigues J, Levitt M and Chopra G†
Nucleic Acid Research, 2012, Web Server Issue. 40 (W1): W323-W328
PMCID: PMC3394243
Summary: The KoBaMIN web server provides an online interface to a simple, consistent and computationally efficient protein structure refinement protocol based on minimization of a knowledge-based potential of mean force.

Remarkable patterns of surface water ordering around polarized buckminsterfullerene
Chopra G† and Levitt M
Proc. Natl. Acad. Sci. U.S.A., 2011, 108(35):14455-14460
PMID: 21844369
Summary: Molecular Dynamics simulations using a state-of-the-art quantum mechanical polarizable force field (QMPFF3) shows that polarization increases ordered water structure so that the imprint of the hydrophobic surface atoms on the surrounding waters is stronger and extends to long-range.

Consistent refinement of submitted models at CASP using a knowledge-based potential
Chopra G†, Kalisman N, Levitt M
Proteins, 2010, 78(12): 2668-2678
PMC2911515

Solvent dramatically affects protein structure refinement
Chopra G, Summa CM, Levitt M
Proc. Natl. Acad. Sci. U.S.A., 2008, 105(51): 20239-20244
PMID: 19073921