CANDOCK:
Computational ANalytics based DOCKing

CANDOCK Overview
CANDOCK is a fast, hierarchical, flexible, and knowledge-based set of docking tools designed to determine the interactions a given small-molecule, nucleic acid, or protein has with an entire proteome. This tool is freely available for academic use.
The CANDOCK software package is written in the C++-14 programing language and is distributed as a library and set of tools linking against this library. Each tool in the software package is discussed below in the 'How it works' section.
How it works
The design of CANDOCK has been carried out in a manner leading to a software package that is easy to use, fast, and accurate. Users of the software need only to give a biological target of interest (protein, or nucleic acid, for example) and the 3D structures of the ligand they are interested in docking.
CANDOCK does not need a binding site to be explicitly given and will predict a binding pocket using the ProBIS algorithm if the user does not specify a pocket. Binding site prediction can be invoked directly using the find_centroids tool. This feature is in contrast to other commercial and academic software which requires the user to provide a binding pocket, simplifying the utilization of this software package.
Fragmentation of ligands is crucial to CANDOCK's speed and accuracy. Molecules of interest are broken down into rigid fragments which possess no single bonds not confined in a ring structure. This process, along with atom typing and other pre-docking steps is carried out by the prep_fragments tool.
The dock_fragments tool places rigid bodies in the binding pocket of interest, then rotates and translates these fragments as to explore the rigid body space each fragment has within the search area. Once all the possible conformations of a fragment are determined, the poses are clustered and scored using a knowledge-based potential and referred to as ligand 'seeds'.
To obtain the final docked pose of the ligand, the maximum clique algorithm takes the seeds generated in the previous step and determines all possible conformations that may occur in the binding pocket of the protein. This algorithm is incredibly fast and can create millions of structures in seconds. Evaluation of each structure is carried out to determine if the geometry of the structure is reasonable, resulting in a few thousand structures. Each structure undergoes an energy minimization procedure and is presented to the user as the initial structure docked in the binding pocket. This process is performed by the link_fragments tool.
Examples
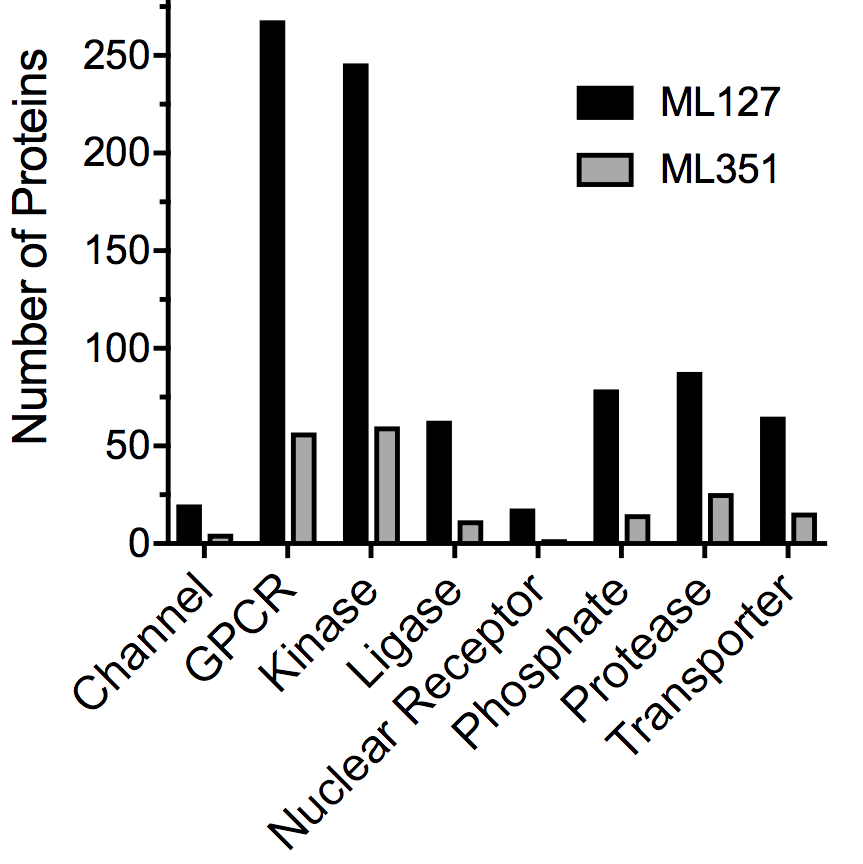
Above are the results of docking two leads (ML127 and ML351) to the entire human proteome. Specific classes of proteins were selected due to their association with toxicity and the number of times each lead hits theses proteins is shown above. This data, generated by CANDOCK can be used to determine the toxicity of compounds in silico.
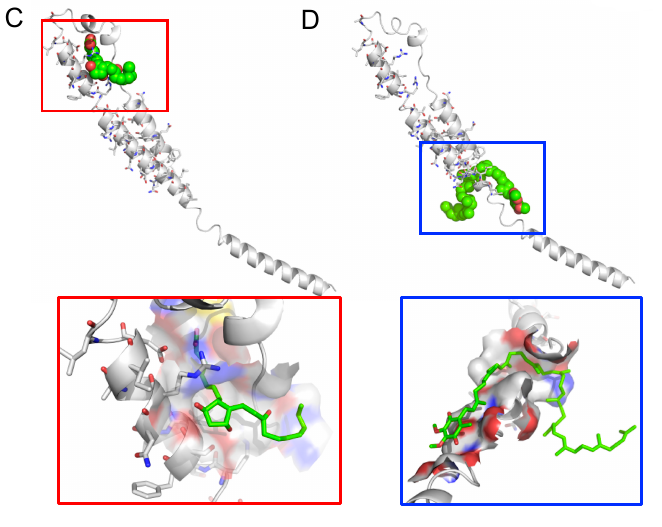
The docking of unoprostone(C) and ubidecarenone(D) to Ebola protein GP2 (PDB identifier 1ebo, chain F) is shown above. This docking was performed flexibly and showcases CANDOCK's ability to dock large molecules to large targets.