|
| |
Our research projects are focused on understanding the mechanisms and pathways through
which the expression of human genes is controlled.
- For studies of the Human Genome, we develop tools
in Bioinformatics
and theoretical models for making
experimentally testable predictions. These efforts are interdisciplinary
projects
done in collaboration with scientists from the Department of Biology, Statistics,
Computer Science and Electrical Engineering.
- For studies of the Human
Genome, we have collected the sequences of the experimentally
defined transcription factor
binding sites. The goal of this project is
to resolve the redundancy problem encountered in the current databases of
transcription factor binding elements.
- We are developing computational
and statistical models to locate the regulatory
regions in human genomic DNA.
- We are constructing a database
of a subset of the human regulatory genes,
including transcription factors. The goal of this project is to
to reconstruct the regulatory networks that
control the expression of human genes.
- We have construced a
database (Rosalind_Franklin.hsDAT) of potential regulatory codes in human
DNA. The goal of this project is to discover "the
linguistics" of DNA defining the regulatory
regions of human genes.
- Use of this database along with contruction of density plots facilitate localization of potential regulatory codes in human genomic DNA
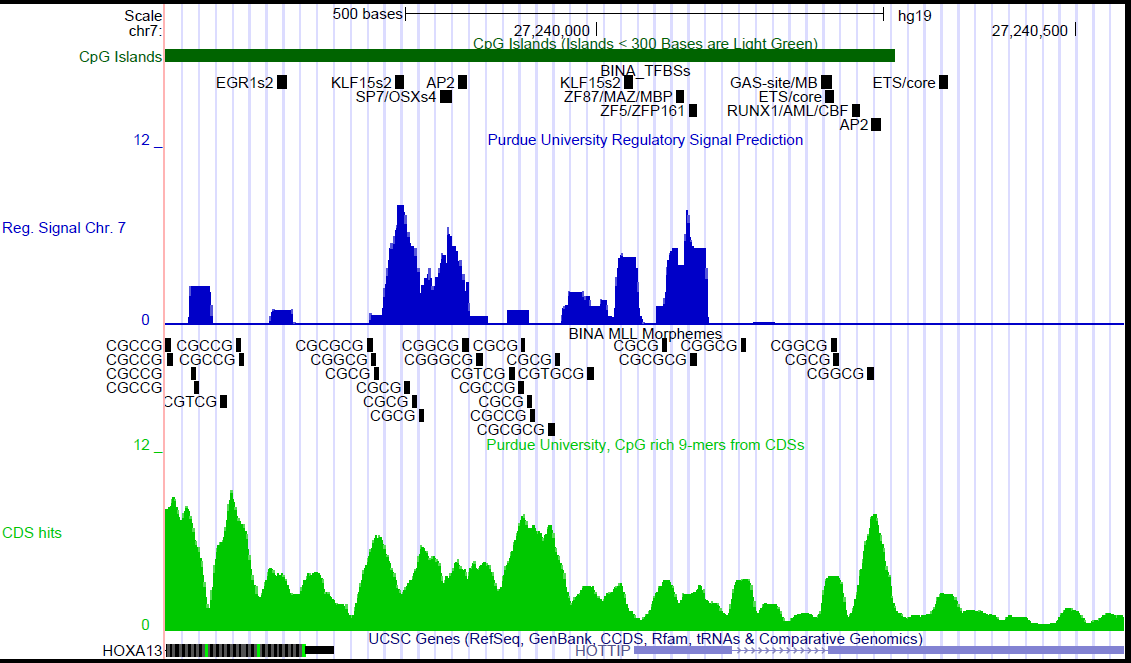
- We have discovered DNA sequence elements (morphemes) that bind MLL. Localization of such may identify the regions that recruit the human trithorax complexes to genomic DNA to maintain active chromatin states during mitosis.
As an example, the diagram below shows the position of potential transcription factor binding sites with respect to computationally defined regulatory codes (blue) and the position of MLL morphemes in the promoter region of the human HOXA13 gene.
|
