We are focusing onProtein ubiquitination and bacterial pathogens
Latest Research
Research area 1
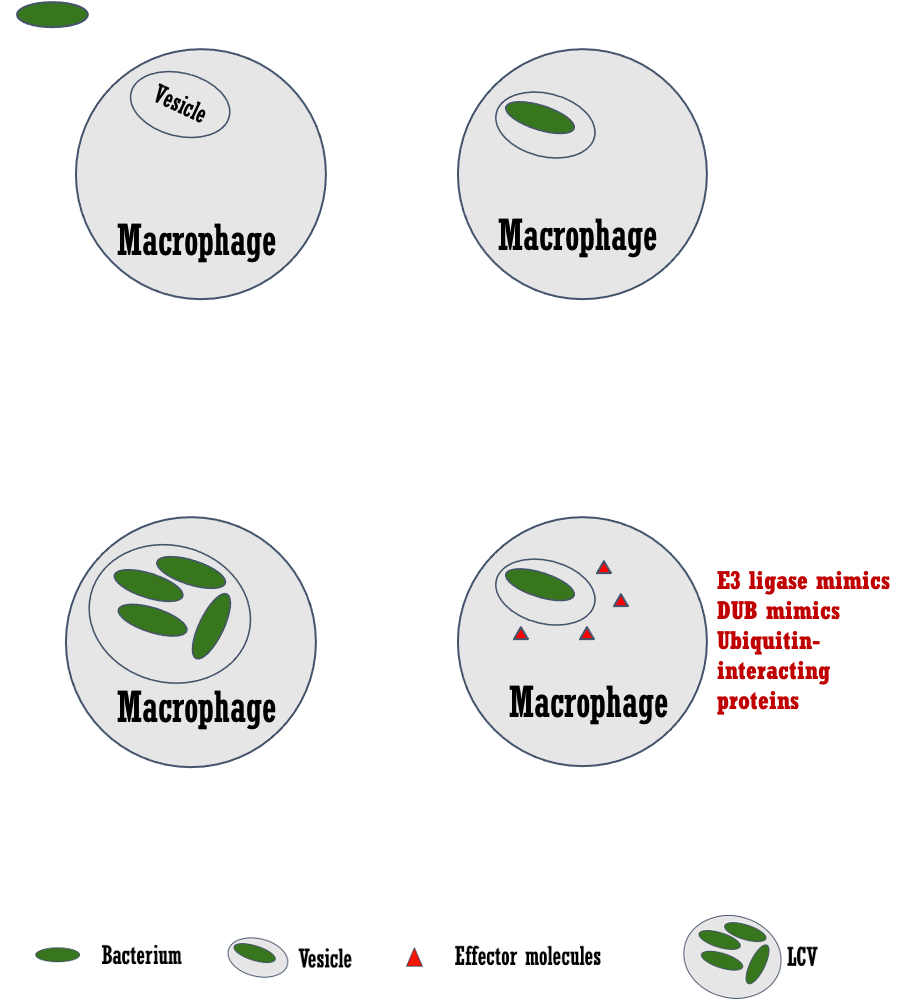
The broad research interest of my group centers around protein ubiquitination and its relationship with bacterial pathogens. Specifically, understanding mechanism of action of deubiquitinases (DUBs) and ubiquitin modifying enzymes of both prokaryotic and eukaryotic origin. Using crystallography, biochemical and biophysical data we wish to unravel mechanism of action of these enzymes, including regulation of catalytic activity and substrate selection. The fact that viruses and bacteria manipulate our ubiquitination pathways has been known for at least two decades, with a number of well characterized examples of pathogens using enzymes or substrate adaptors to co-opt our ubiquitination pathways.
Research area 2
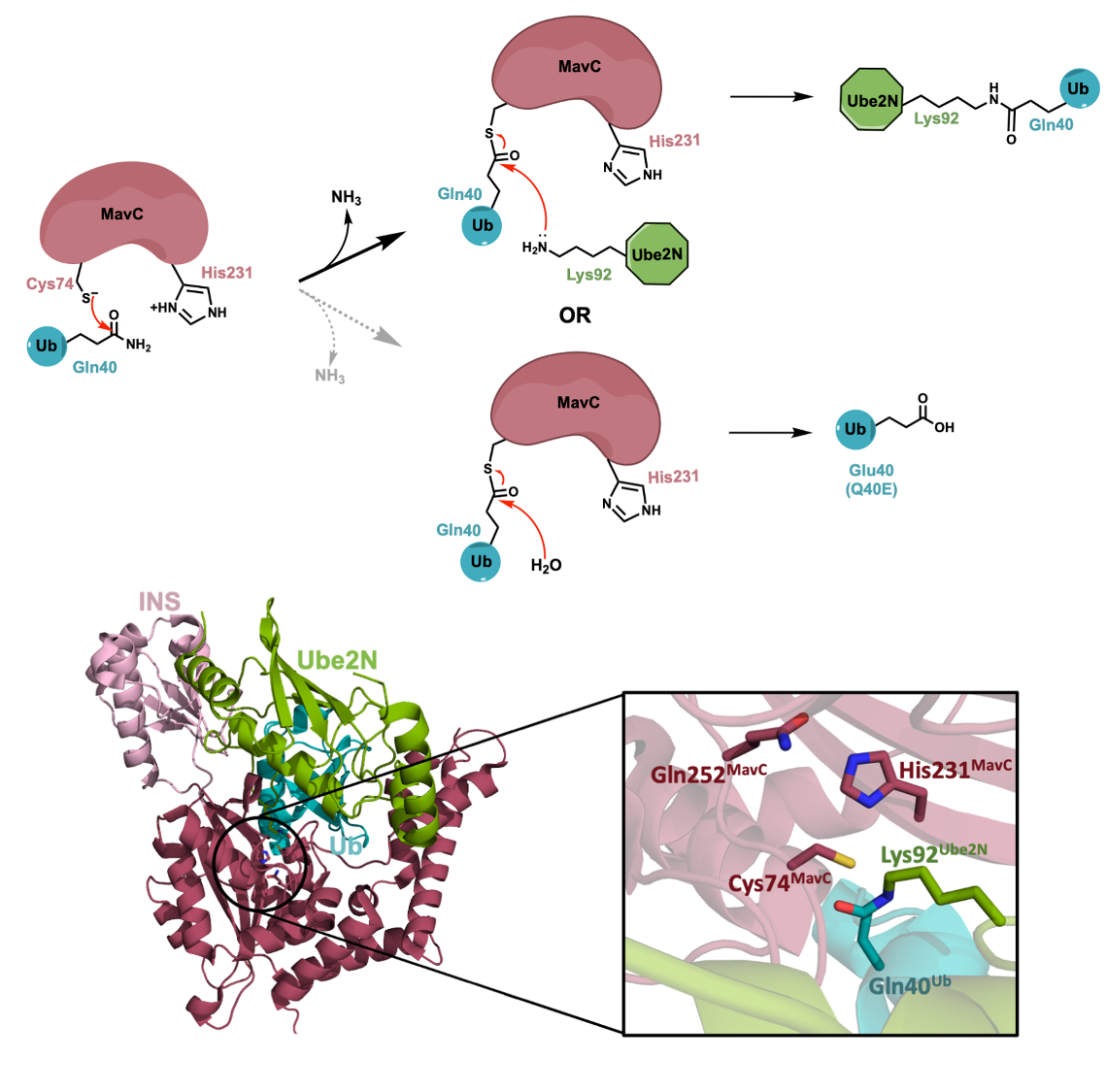
We are engaged in dissecting the mechanism of these enzymes, specifically, how their catalytic activities are regulated. The enzymes we are studying are known to be activated when they associate with larger macromolecular complexes. We use X-ray crystallography in conjunction with a variety of structural, biochemical, and biophysical techniques to accomplish these goals.

Research area 3
We are engaged in dissecting the mechanism of these enzymes, specifically, how their catalytic activities are regulated. The enzymes we are studying are known to be activated when they associate with larger macromolecular complexes. We use X-ray crystallography in conjunction with a variety of structural, biochemical, and biophysical techniques to accomplish these goals.

Research area 4
We are engaged in dissecting the mechanism of these enzymes, specifically, how their catalytic activities are regulated. The enzymes we are studying are known to be activated when they associate with larger macromolecular complexes. We use X-ray crystallography in conjunction with a variety of structural, biochemical, and biophysical techniques to accomplish these goals.