Research
Regulation of ubiquitin signaling in humans
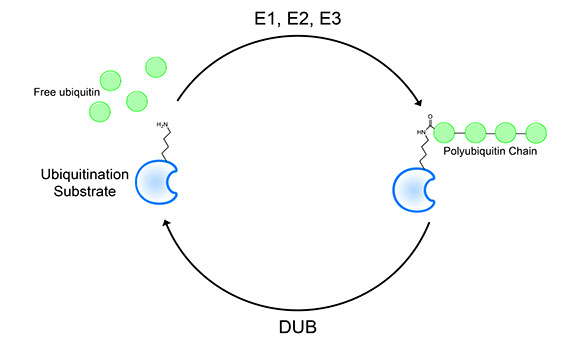
Ubiquitin is a small protein first discovered in 1975. Through further studies of this protein, it was discovered that this 8.5 kDa protein is a major player in many signaling pathways in living organisms. In the human system, ubiquitination is done by the assistance of three major types of enzymes: E1, E2, and E3.
E1 has a function to activate the ubiquitin, rendering it ready to be attached to the E2 enzyme. The E2 enzyme is the ubiquitin-conjugating enzyme that will be brought to the E3 enzyme. Finally, the E3 enzyme is the enzyme that transfers the ubiquitin on E2 to the substrate protein. This makes the specificity to be highly determined by the E3 complex. Deubiquitinases are proteins that catalyze the removal of these ubiquitin linkages to replenish the cellular ubiquitin pool.Non-canonical Ubiquitination by Pathogenic Effectors
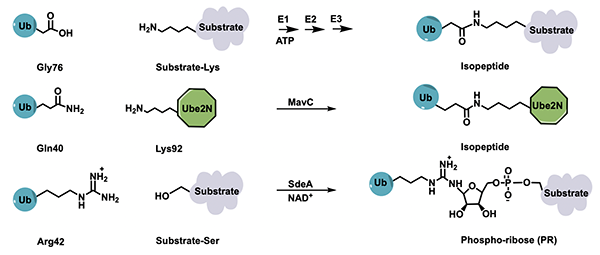
My group has played an instrumental role in identifying key aspects of the first ever known ubiquitination mechanism outside the eukaryotic E1-E2-E3 system, a discovery that stemmed from our identification of a novel deubiquitinating enzyme in a bacterial pathogen. More recently, we have been able to provide structural basis of another novel ubiquitination reaction that makes use of a transglutaminase chemistry for crosslinking ubiquitin to its target. These exciting results indicate that we have underestimated the scope of bacterial interference in our ubiquitination pathways; it is quite possible that more new mechanisms exist awaiting discovery. We are excited about this possibility and are looking into the effector arsenal of a number of intracellular pathogens for new ubiquitin-interacting effectors in the hopes of identifying novel aspects of host-pathogen interaction.
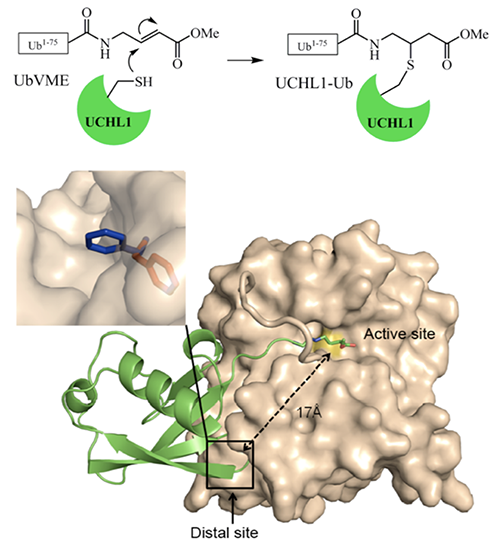
Techniques used include X-ray crystallography, cryo-EM, mass spectrometry, SDS-PAGE gel-based assays, and fluorescence spectroscopy.Ubiquitin-based probe design for covalent linkage
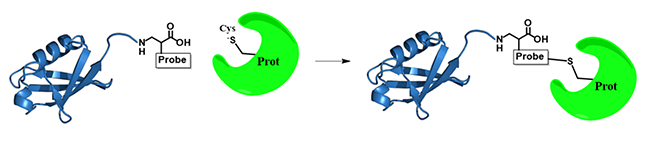
Our lab develops ubiquitin-based probes as a way to identify ubiquitin binding proteins in the cells. We create mutants on different ubiquitin residues that will incite covalent linkages formation, either by the use of disulfide bonds, unnatural amino acid incorporation, or electrophilic suicide inhibitor. We perform this analysis between ubiquitin and different types of ubiquitin binding enzymes, mainly deubiquitinating enzymes and pathogenic effector proteins. We aim to better characterize the specific binding site of ubiquitin in these enzymes. Additionally, this project looks to take this probe design to covalently link ubiquitin binding protein for proteomics.
We employ our molecular biology toolbox to incorporate this probes to bring it to biochemical assays, x-ray crystallography, and proteomics.
Identification and characterization of ubiquitin-interacting effectors in Legionella pneumophila
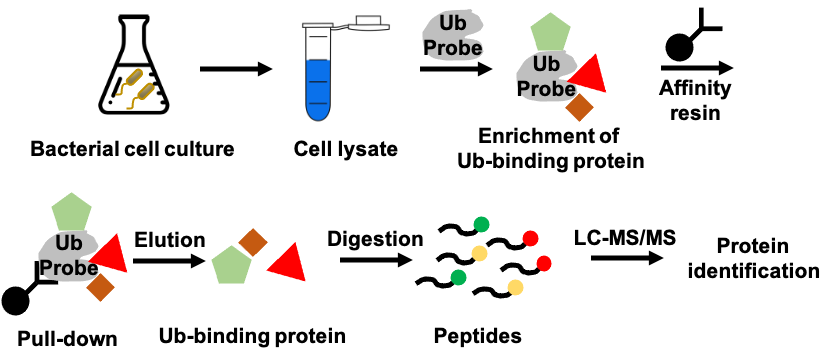
Secreted protein effectors contribute significantly to the pathogenesis of Legionella pneumophila and interfere intensively with host ubiquitin signaling pathway, which is crucial for host immune response and protein degradation. Investigation on how these effectors interact with ubiquitin system can help us with understanding the mechanism of Legionella infection and subsequent drug discovery process. To this end, we utilize activity-based ubiquitin probes to capture ubiquitin-interacting effectors and characterize their functions to uncover their relations to ubiquitin signaling pathway.
Techniques used in this project include activity-based proteomics, X-ray crystallography, biochemical and biophysical assays complemented by cell biology experiments.
Chemical biology of ubiquitin signaling
Thanks to the collective effort of a large number of scientists spanning the past three decades or so, many basic aspects of ubiquitination reaction and a general understanding of its cellular function have become relatively well established. However, an understanding of how ubiquitin signaling works in the context of tissue-specific functions is still in its infancy. In this broad aim, we have undertaken at least two collaborative projects with the hopes of shedding some light into how ubiquitin signaling works tissue specific function such as in immune cells and neurons. Toward these goals, we think a better approach would be use chemical and genetic probes to understand the underlying biology. The chemical probes are being developed starting from small molecule inhibitors through fragment screening approach, which is being led by the Flaherty group (MCMP, Purdue University). We also are collaborating groups with expertise in chemical biology to better understand tissue specific function of protein ubiquitination.